Natural Bond Orbital.
Natural Bond Orbitals (NBOs) are localized few-center orbitals ("few" meaning typically 1 or 2, but occasionally more) that describe the Lewis-like molecular bonding pattern of electron pairs (or of individual electrons in the open-shell case) in optimally compact form. More precisely, NBOs are an orthonormal set of localized "maximum occupancy" orbitals whose leading N/2 members (or N members in the open-shell case) give the most accurate possible Lewis-like description of the total N-electron density.
Neither the form of the bonding hybrids nor the locations of localized bonds and lone pairs are pre-determined. Rather, the NBO program searches over all possible ways of drawing the bonds and lone pairs for the variationally optimal bonding pattern that places maximum occupancy (highest percentage of the total electron density) in the leading N/2 "Lewis-type" NBOs (typically >99.9% for common organic molecules). The Lewis-type NBOs determine the localized Natural Lewis Structure (NLS) representation of the wavefunction, while the remaining "non-Lewis"-type NBOs complete the span of the basis and describe the residual "delocalization effects" (i.e., departures from a single localized Lewis structure). Thus, NBOs provide a valence bond-type description of the wavefunction, closely linked to classical Lewis structure concepts. As in the NAO case, the only input to the NBO algorithms is the molecular wavefunction Ψ (through its first-order reduced density operator Γ), so the numerically determined Lewis structure representation is "natural" to Ψ itself.
|
 |
Ethane NBO: Lets found the geometry with the lowest energy.
- First we have to build a geometry which is close to the minimal energy structure.
- After that, we have to make an input file for ORCA program specifying the next:
- ! Opt --> To perform a geometry optimization
- The input file must lool like this ethane.inp
- Copy the orca-3.0.0.pbs file to your work directory, modify the input name and run
user@machine$ cp /share/apps/PBS_scripts/orca-3.0.0.pbs .
user@machine$ qsub orca-3.0.0.pbs
After all this, you have the optimized geometry at the end of the *.trj file. |
Figure 1. Ethane
|
Generating the file *.47 with orca
Once we have our optimized geometry, we use it to generate the input file for NBO6.0
- Create a new folder and copy to it the next files, *.inp the orca3.0.0.pbs.
- Open the input file, delete the !Opt instruction and add this new one !NPA
- Delete the old geometry
user@machine$ mkdir nbo
user@machine$ cp file.inp nbo
user@machine$ cp orca-3.0.0.pbs nbo
user@machine$ vi file.inp
Input file must look like ethane_nbo.inp. After modification again run Orca
user@machine$ qsub orca-3.0.0.pbs
The NPA instructions will allow us to generate the input file for NBO6.0 the file *.47
|
Figure 2. Input example
|
Using the orca_nbo subprogram
In order to use the orca_nbo routine we must change a few lines on the *.47 file. Please open the file *.47 and follow the instructions below.
- Remove the SKIPNBO keyword
- Add PLOT
- Add NBO
After this, you have to excecute:
user@machine$ orca_nbo &
This instruction will return several files which can be view from a plotting program like ChemCraft |
Figura 3. NPA input file
|
Plotting NBO with Chemcraft.
Chemcraft is a powerfull tool to visualize outputs from quantum chemical softwares, an also to plot a wide range of molecular properties, it can be found at www.chemcraftprog.com.
If you are on a server you first have to download the working directory to a known directory on your local hard drive, if you are on the same computer were you perform the calculus you can skip this step.
|
  |
Let's visualize ethane molecule.
- Execute the ChemCraft program.
- Open the file *.31
This will show us the ethane molecule.
To plot a NBO one has to do the next.
- On the tool bar located at the left choose the FILE37 (NBO)
- Then at the bottom of the same tool bar click on tools>render molecular orbitals
- Choose the molecular orbitlas (MO's) you want to visualize. in this case we select until the 13.
- After this, all selected MO's appeare on the left tool bar.
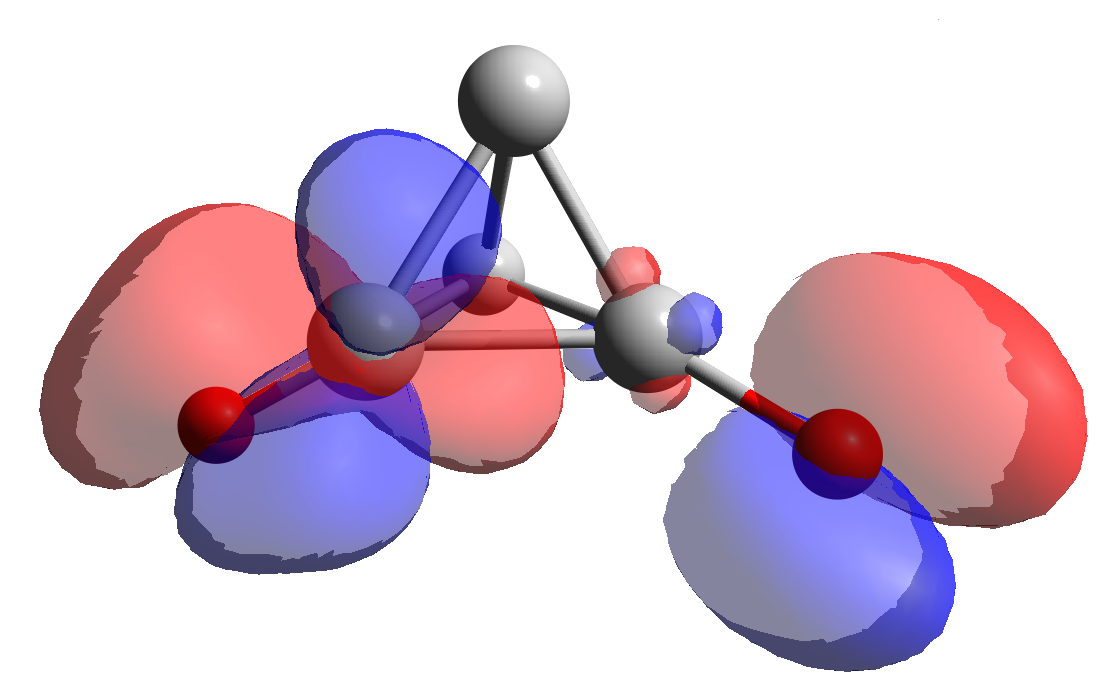
- Select the ninth MO, it appears a new tool menu, click show isosurface and both signed it must look like Figure 4. |
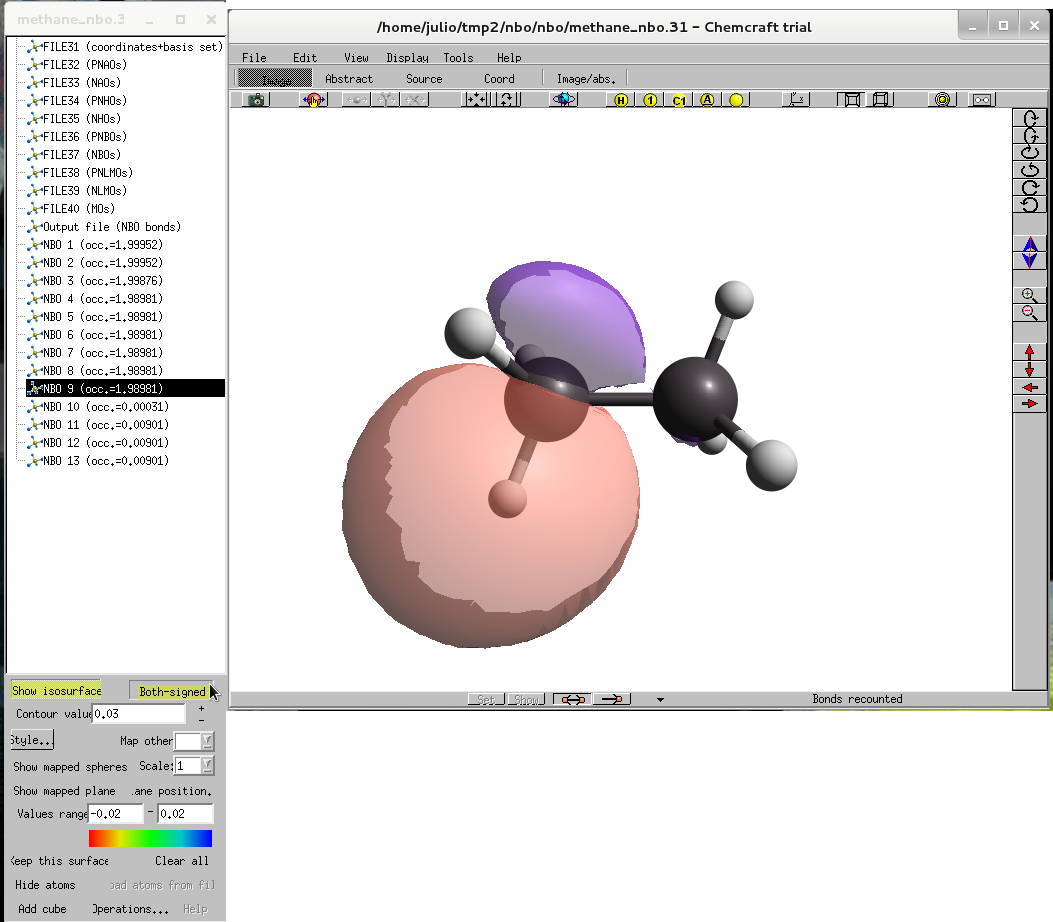
Figure 4. HOMO of ethane. |